Science & Technology
Whole Genome Sequencing
- 19 Dec 2022
- 9 min read
For Prelims: Whole Genome Sequencing, DNA, Gene, Genome.
For Mains: Whole Genome Sequence and its Significance
Why in News?
Recently, Researchers at the Indian Institute of Science Education and Research (IISER) Bhopal have carried out Whole Genome Sequencing of banyan (Ficus benghalensis) and peepal (Ficus religiosa) from leaf tissue samples.
- The work helped in identifying 17 genes in the case of banyan and 19 genes of peepal with multiple signs of adaptive evolution (MSA) that play a pivotal role in long-time survival of these two Ficus species.
What is Whole Genome Sequencing?
- About:
- All organisms have a unique genetic code, or genome, that is composed of nucleotide bases- Adenine (A), Thymine (T), Cytosine (C) and Guanine (G).
- The unique Deoxyribonucleic Acid (DNA) fingerprint, or pattern can be identified by knowing the sequence of the bases in an organism.
- Determining the order of bases is called sequencing.
- Whole genome sequencing is a laboratory procedure that determines the order of bases in the genome of an organism in one process.
- Methodology:
- DNA Shearing:
- Scientists begin by using molecular scissors to cut the DNA, which is composed of millions of bases (A’s, C’s, T’s and G’s), into pieces that are small enough for the sequencing machine to read.
- DNA Bar Coding:
- Scientists add small pieces of DNA tags, or bar codes, to identify which piece of sheared DNA belongs to which bacteria.
- This is similar to how a bar code identifies a product at a grocery store.
- Scientists add small pieces of DNA tags, or bar codes, to identify which piece of sheared DNA belongs to which bacteria.
- DNA Sequencing:
- The bar-coded DNA from multiple bacteria is combined and put in a DNA sequencer.
- The sequencer identifies the A’s, C’s, T’s, and G’s, or bases, that make up each bacterial sequence.
- The sequencer uses the bar code to keep track of which bases belong to which bacteria.
- Data Analysis:
- Scientists use computer analysis tools to compare sequences from multiple bacteria and identify differences.
- The number of differences can tell the scientists how closely related the bacteria are, and how likely it is that they are part of the same outbreak.
- DNA Shearing:
- Advantages:
- Provides a high-resolution, base-by-base view of the genome
- Captures both large and small variants that might be missed with targeted approaches
- Identifies potential causative variants for further follow-up studies of gene expression and regulation mechanisms
- Delivers large volumes of data in a short amount of time to support assembly of novel genomes
- Significance:
- Genomic information has been instrumental in identifying inherited disorders, characterizing the mutations that drive cancer progression, and tracking disease outbreaks.
- It is beneficial for sequencing agriculturally important livestock, plants, or disease-related microbes.
What is Genome?
- A genome refers to all of the genetic material in an organism, and the human genome is mostly the same in all people, but a very small part of the DNA does vary between one individual and another.
- Every organism’s genetic code is contained in its DNA, the building blocks of life.
- The discovery that DNA is structured as a “double helix” by James Watson and Francis Crick in 1953, started the quest for understanding how genes dictate life, its traits, and what causes diseases.
- Each genome contains all of the information needed to build and maintain that organism.
- In humans, a copy of the entire genome contains more than 3 billion DNA base pairs.
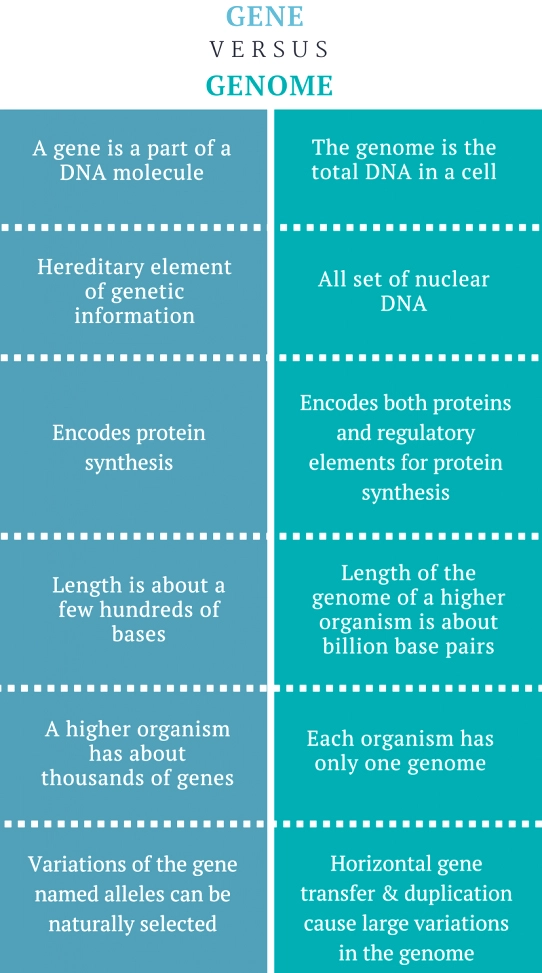
What is the Difference between Genome and Gene?
UPSC Civil Services Examination, Previous Year Questions (PYQs)
Q1. With reference to agriculture in India, how can the technique of ‘genome sequencing’, often seen in the news, be used in the immediate future? (2017)
- Genome sequencing can be used to identify genetic markers for disease resistance and drought tolerance in various crop plants.
- This technique helps in reducing the time required to develop new varieties of crop plants.
- It can be used to decipher the host-pathogen relationships in crops.
Select the correct answer using the code given below:
(a) 1 only
(b) 2 and 3 only
(c) 1 and 3 only
(d) 1, 2 and 3
Ans: (d)
Exp:
- Chinese scientists decoded rice genome in 2002. The Indian Agricultural Research Institute (IARI) scientists used the genome sequencing to develop better varieties of rice such as Pusa Basmati-1 and Pusa Basmati-1121, which currently makes up substantially in India’s rice export. Several transgenic varieties have also been developed, including insect resistant cotton, herbicide tolerant soybean, and virus resistant papaya. Hence, 1 is correct.
- In conventional breeding, plant breeders scrutinize their fields and search for individual plants that exhibit desirable traits. These traits arise spontaneously through a process called mutation, but the natural rate of mutation is very slow and unreliable to produce all the plant traits that breeders would like to see. However, in genome sequencing it takes less time, thus it is more preferable. Hence, 2 is correct.
- The host-pathogen interaction is defined as how microbes or viruses sustain themselves within host organisms on a molecular, cellular, organism or population level. The genome sequencing enables the study of the entire DNA sequence of a crop, thus it aids in understanding of pathogens’ survival or breeding zone. Hence, 3 is correct.
- Therefore, option (d) is the correct answer.
Q2. Consider the following statements: (2022)
DNA Barcoding can be a tool to:
- assess the age of a plant or animal.
- distinguish among species that look alike.
- identify undesirable animal or plant materials in processed foods.
Which of the statements given above is/are correct?
(a) 1 only
(b) 3 only
(c) 1 and 2
(d) 2 and 3
Ans: (d)
Exp:
- The novel technique of identifying biological specimens using short DNA sequences from either nuclear or organelle genomes is called DNA barcoding.
- DNA barcoding has many applications in various fields like preserving natural resources, protecting endangered species, controlling agriculture pests, identifying disease vectors, monitoring water quality, authentication of natural health products and identification of medicinal plants.
- Species identification of endangered wildlife (hence, distinguishes among species that look alike), quarantine pests, and disease vectors (identifying undesirable animals/plants) are just a few areas in which DNA barcoding is enabling researchers, enforcement agents, and consumers to make informed decisions in much shorter time frames.
- Statement 2 and statement 3 are correct, Hence, option (d) is correct (by elimination).
Mains
Q. What are the research and developmental achievements in applied biotechnology? How will these achievements help to uplift the poorer sections of society? (2021)